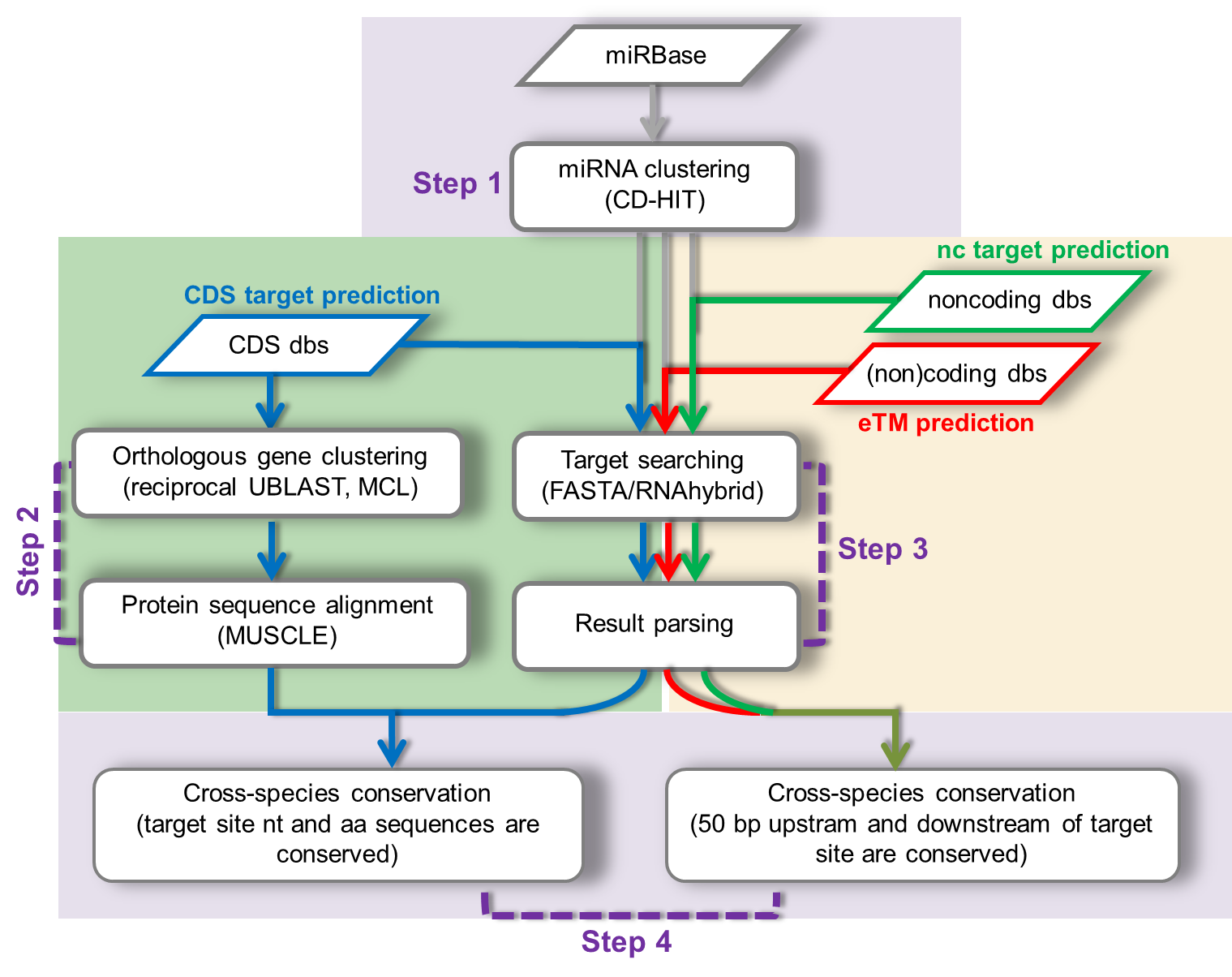
TarHunter predicts conserved miRNA targets and endogenous target mimics (eTM) in plants. It uses a strict cross-species conservation filter that resembles TargetScan, an animal miRNA target predictor, and predicts eTM based on the criterion emphasizing central mismatch/bulge rather than a bulge.
TarHunter consists of three sections: coding sequence (CDS) target prediction, noncoding target prediction and eTM prediction, as illustrated in the following graph.
This website collects TarHunter predictions, degradome analysis results and predicted noncanonical miRNA targets from a variety of plant species. To use TarHunter, please cite the following paper.
Ma X. et al. TarHunter, a tool for predicting conserved microRNA targets and target mimics in plants. Bioinformatics. 2018;34(9):1574-1576.
We recently created TarDB, a plant miRNA target and miRNA-triggered phasiRNA database.
![]()
Copyright @ 2016 Contact: Dr. Xuan Ma
College of Life Sciences,Tianjin Normal University,Tianjin, 300387, China