Brief Introduction
About TarDB
TarDB is a plant miRNA target repository, which is based on cross-species conservation filter, degradome/PARE-seq, and small RNA-seq data analysis. TarDB collects previously unreported miRNA target and phasiRNA data, most of which are relevant to lineage-specific or species-specific miRNAs. TarDB provides browsing and searching functionalities that allow users to explore miRNA targets in plants.
The conservation filter, degadome/PARE-seq and sRNA-seq analysis provide effective ways to obtain high-confidence miRNA targets. Read More

home
Brief introduction of TarDB. Description of three filters implemented to obtain relatively high-confidence miRNA targets.
browse
Get miRNA targets by browsing plant species, miRNA families or choosing different types of targets.
search
Query three main types of miRNA targets deposited in TarDB by different ways.
download
Download miRNA target data in various plant species.
guide
Guidance of TarDB interfaces, particularly browsing and searching TarDB. Related links and FAQs are also included.
contact
Our contact details. Please email us if you have any comments or suggestions.
43
Plant Species
62888
Conserved miRNA Targets
4304
Degradome-supported Targets
2613
miRNA-triggered PhasiRNA loci
TarDB Plants
TarDB collects 43 species including 24 dicotyledon and 12 monocotyledon plants, 1 basal angiosperm, 1 gymnosperm, 3 Bryophytes and 2 algae species.
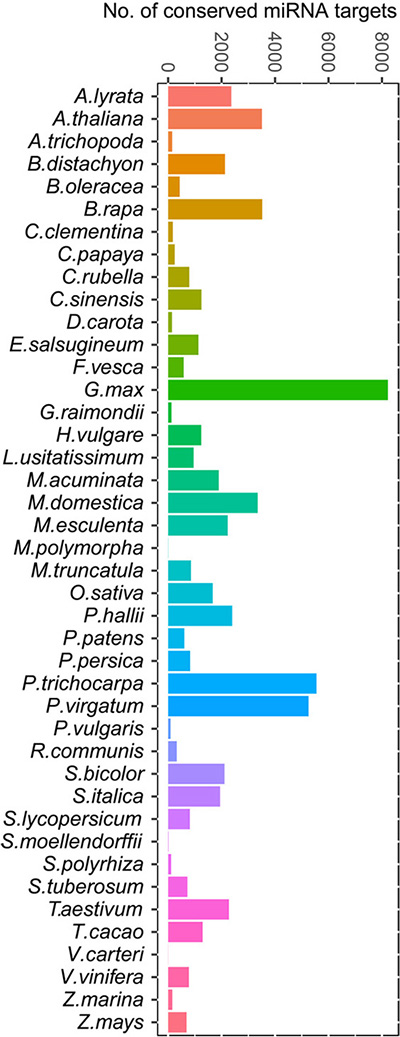
Cross-species Conserved miRNA Targets
TarDB collects 62888 conserved miRNA targets. The concept of TarHunter analysis is conserved miRNAs target homologous genes. See Guide page for details. See more statistics
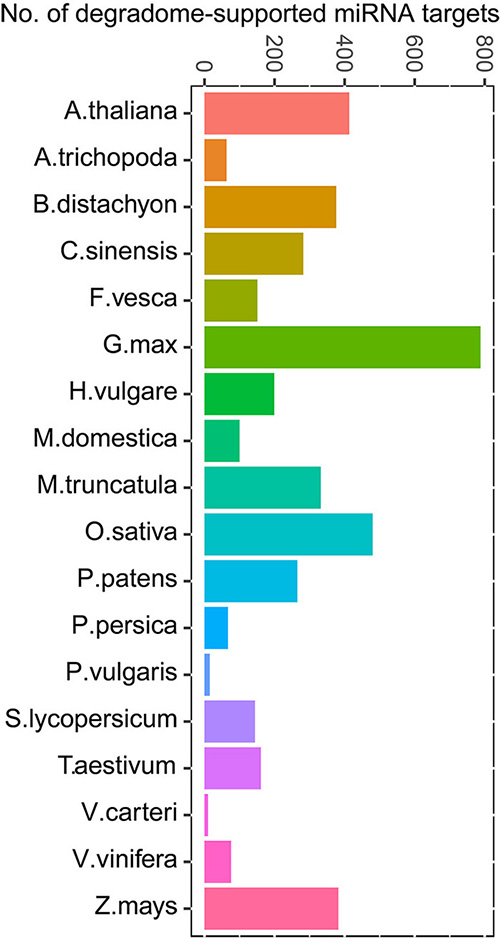
Degradome/PARE-seq Supported miRNA Targets
Degradome analysis identifies 4304 high-confidence miRNA targets deposited in TarDB. See Guide page for details of the criteria in CleaveLand4 analysis. See more statistics
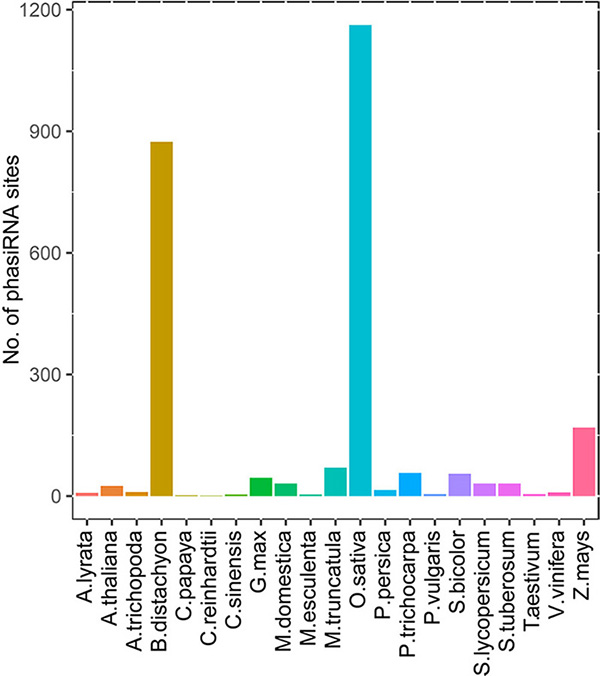
miRNA-triggered PhasiRNAs
TarDB collects 2275 21-nt miRNA-triggered phasiRNA loci from 21 species, and 338 24-nt phasiRNA loci from 10 species. See more statistics
Citation
- Liu J, et al. TarDB: an online database for plant miRNA targets and miRNA-triggered phased siRNAs. BMC Genomics. 2021; 22(1):348.
PMID: 33985427