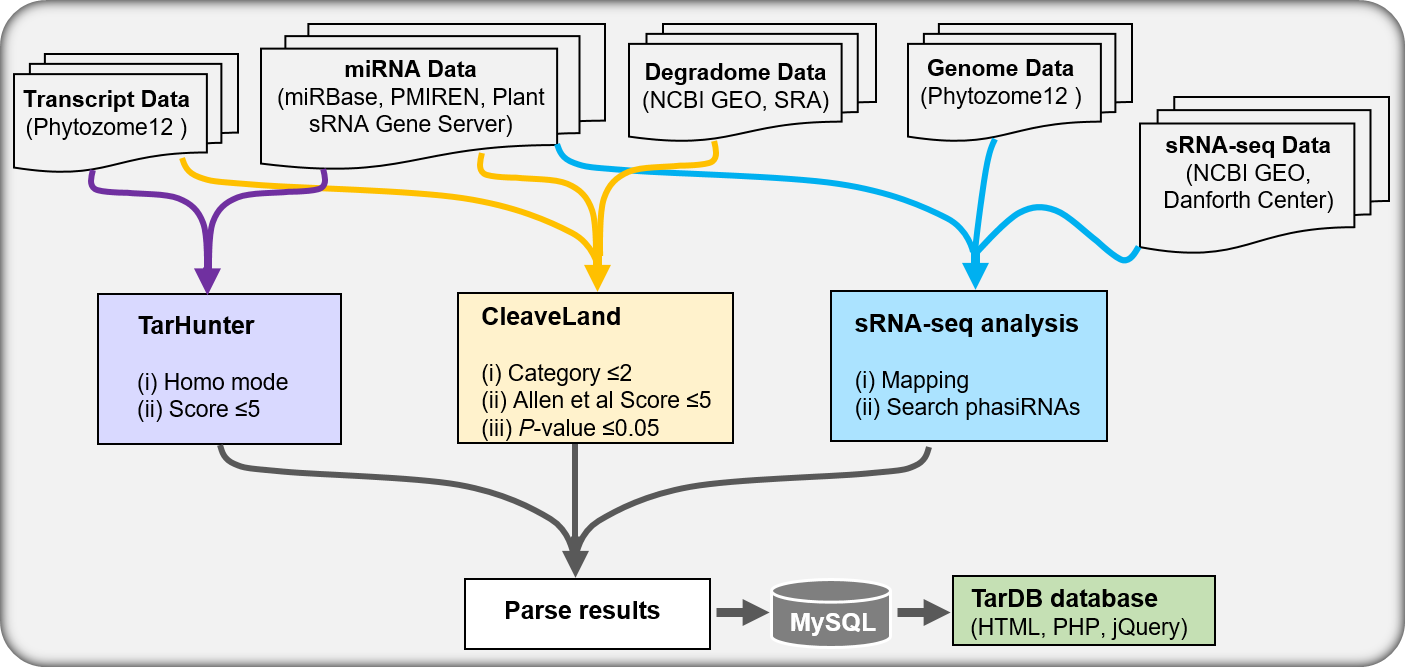
- Cross-species conservation filter is a classic filter in both plant and animal miRNA target predictions.
- Degradome/PARE-seq data provides experimental evidence for identifying miRNA cleaved targets.
- sRNA-seq data facilitate identifying miRNA-triggered phasiRNAs in plants.
TarDB Overview
Welcome to TarDB
In plants, miRNAs regulate target RNAs at posttranscriptional level and occurs in highly sequence complementary manner (Rhoades et al. Cell,2002). This renders plant miRNA target in silico prediction to be relatively easy. Computational programs and web servers have been developed to predict plant miRNA targets, such as TargetFinder, psRNATarget, RNAhybrid and psRobot. However, to obtain relatively high-confidence targets, effective filters are still needed. We previously reported TarHunter, a tool for searching cross-species conserved miRNA targets in plants (Ma et al. Bioinformatics, 2018). Degradome-seq/PARE-seq is currently a robust way for identifying miRNA targets (German et al. Nat Biotechnol,2008; Addo-Quaye et al. Curr Biol,2008). Additionally, small RNA-seq data can be used to identify miRNA triggered phasiRNAs (Allen et al. Cell, 2005; Liu et al. Plant Cell, 2020). To this end, we have integrated conserved miRNA target prediction, degradome/PARE-seq analysis and miRNA-triggered phasiRNA analysis to identify relatively high-confidence miRNA targets. We have created TarDB, an online database that collects different types of miRNA targets and may serve as a useful resource for promoting future studies in plant sRNA field.
Filters
TarDB Implement Three Filters
Workflow
Web interface
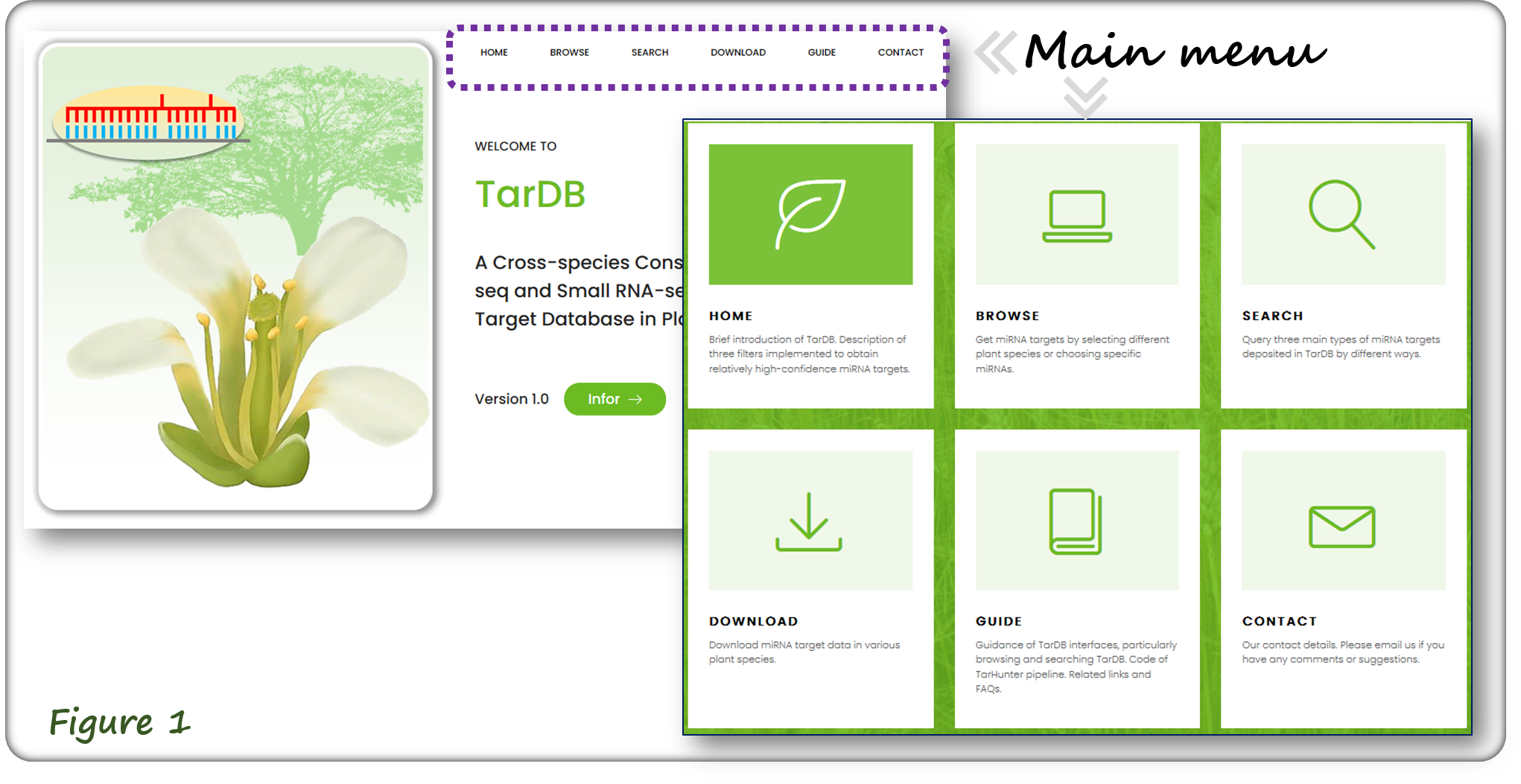
1. Navigation Menu
The main menu at Home page contains six major sections (Figure 1). The Home page presents an overview of miRNA target regulations in plants, and also contains the statistics of miRNA target data in TarDB.
2. Browse
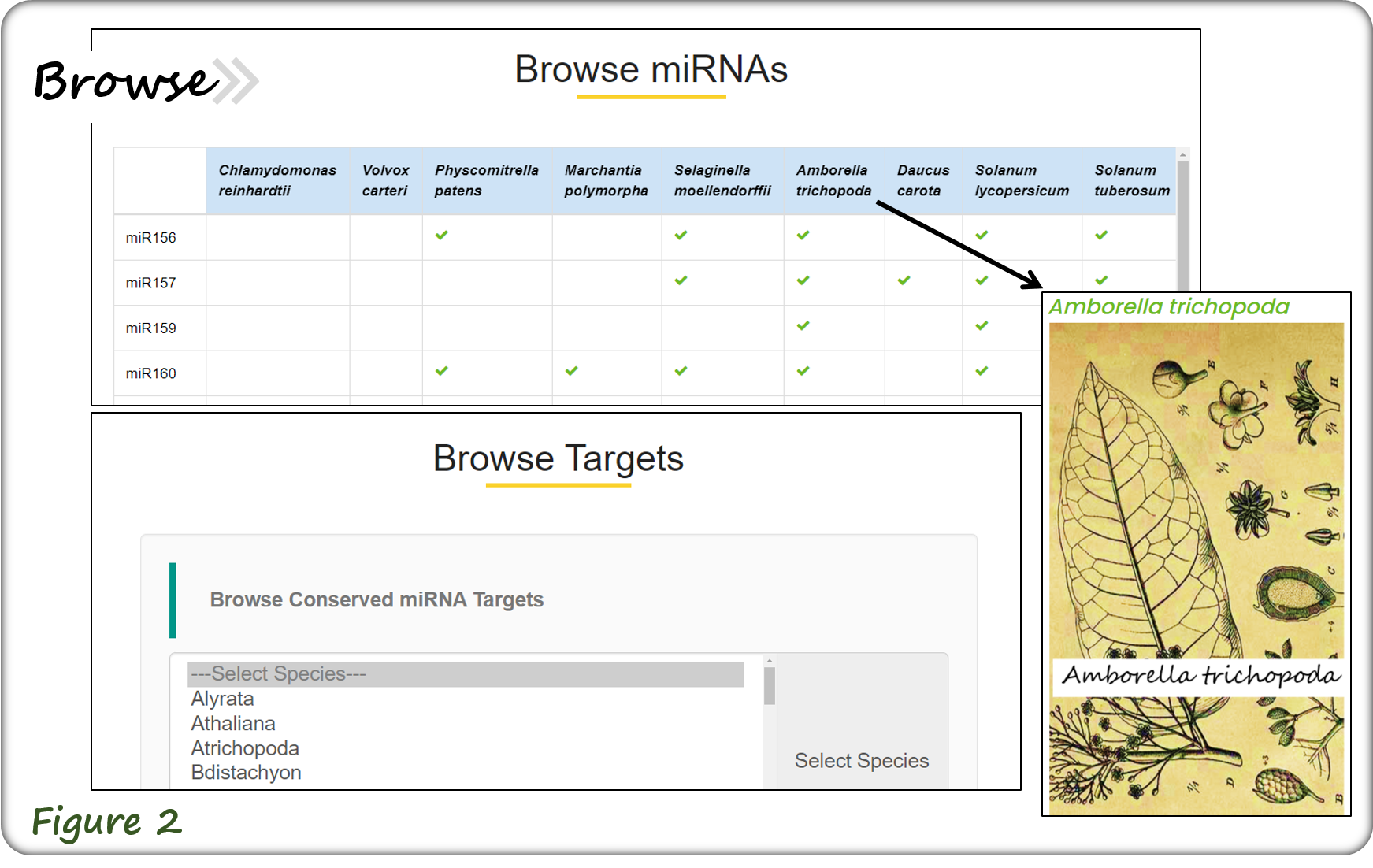
The Browse webpage allows browsing different miRNA families, various plant species and three main types of miRNA targets (Figure 2).
3. Search
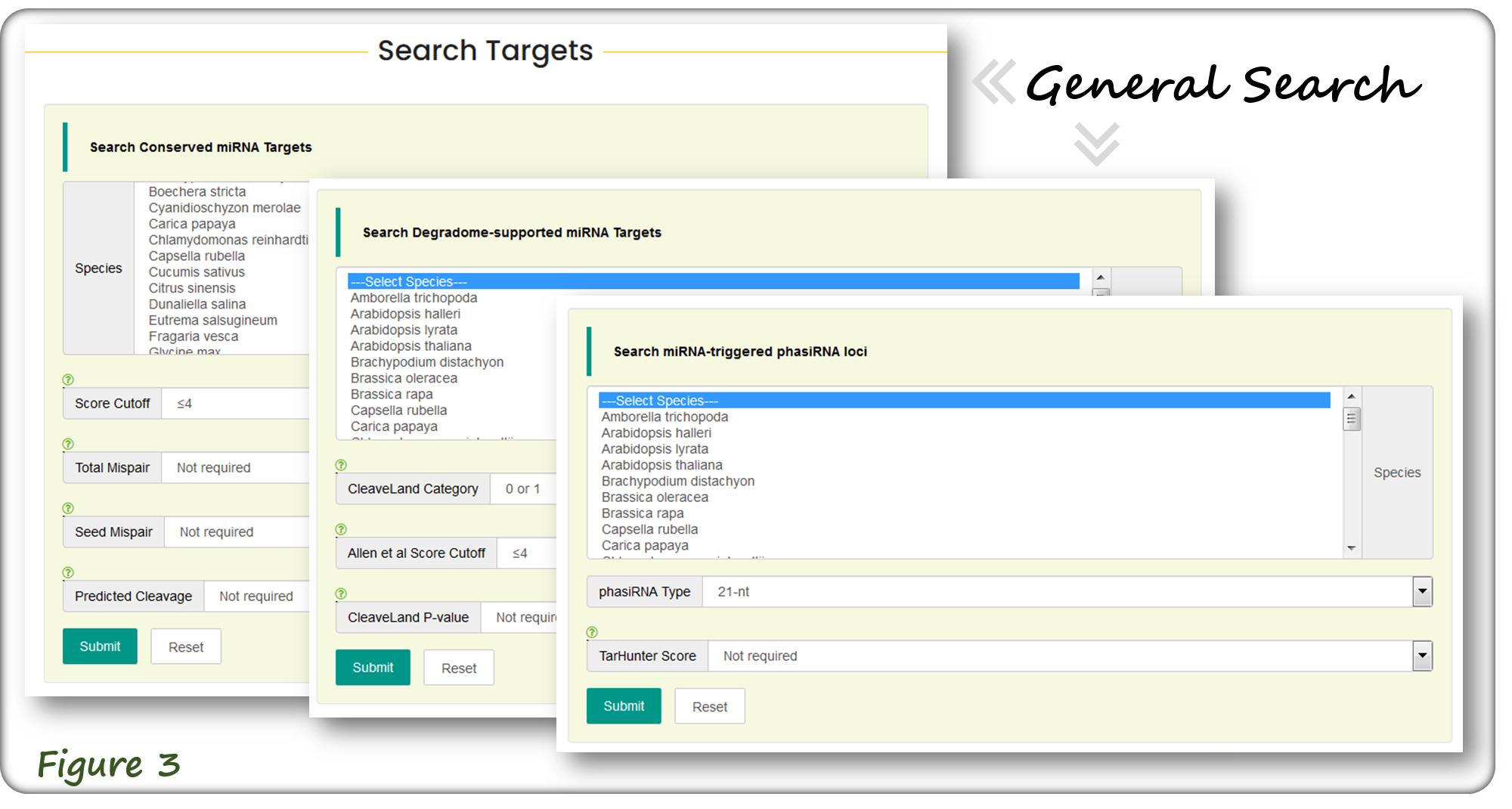
The Search webpage is the most major section of TarDB, which allows for searching conserved miRNA targets, degradome-supported targets and miRNA-triggered phasiRNA loci.
Users can search three types of miRNA targets by general search mode (Figure 3). Alternatively, users can search miRNA targets at a specific genomic locus or by using keywords (Figure 4).
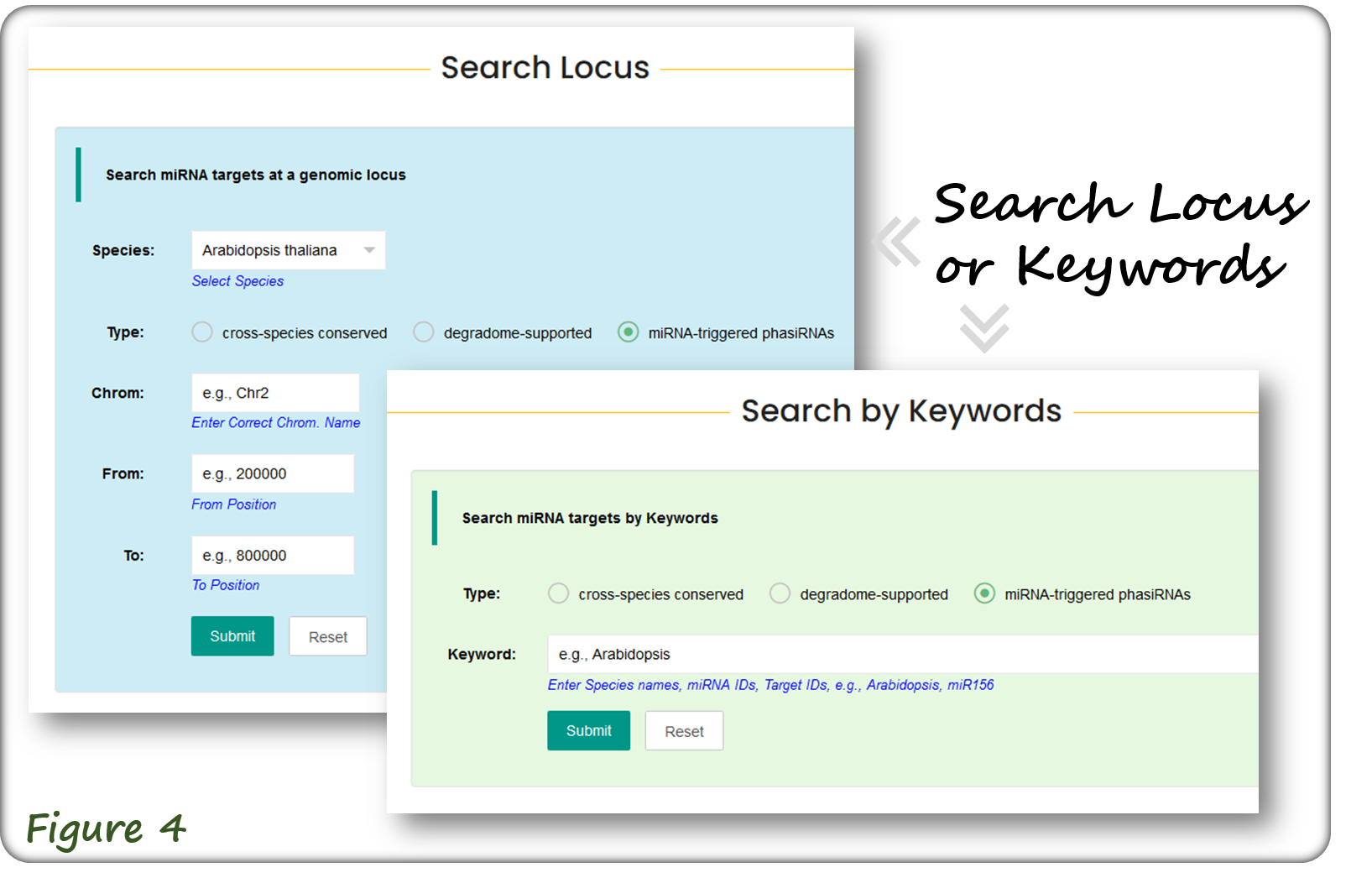
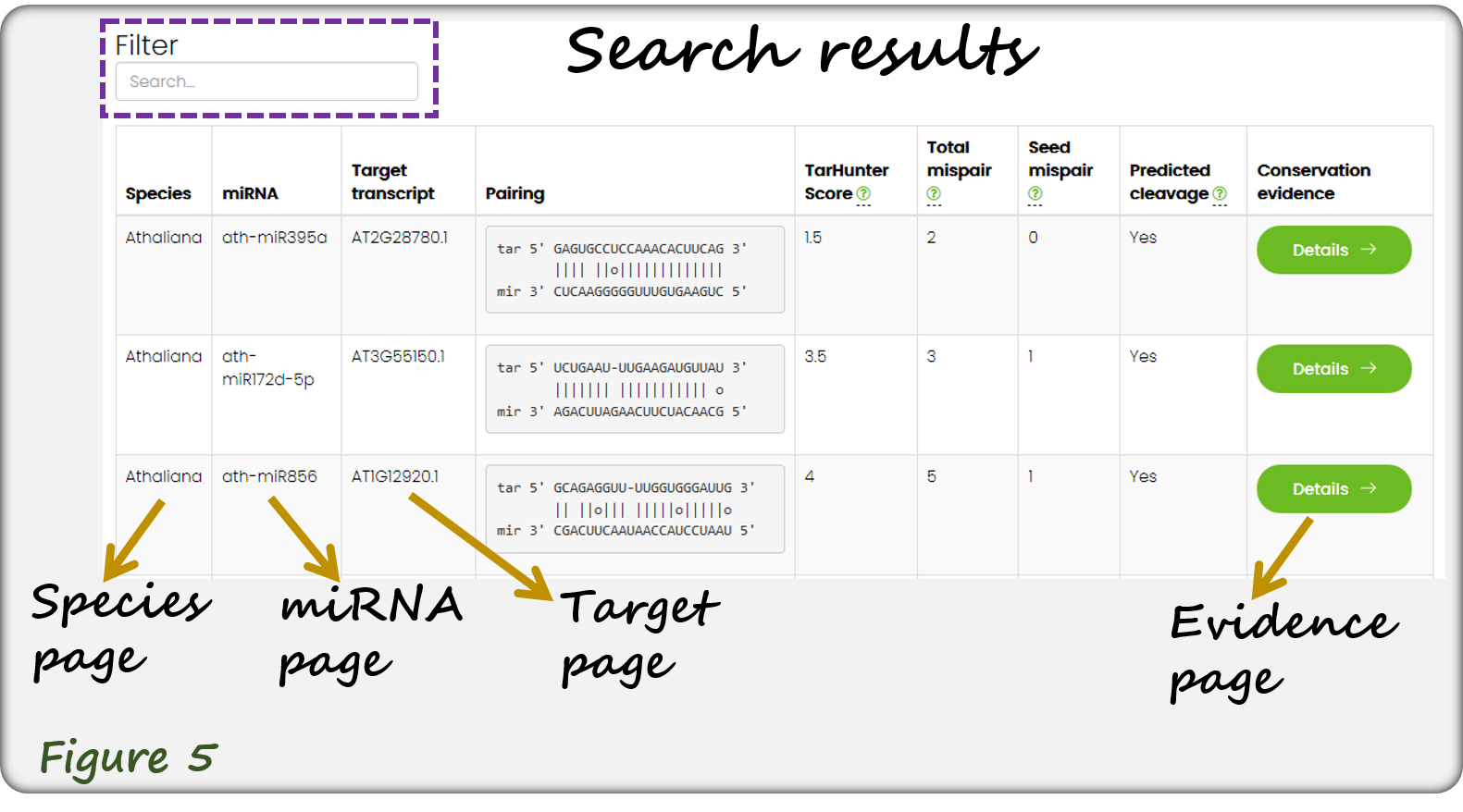
The query results are displayed in tabular format as shown below(Figure 5). Users can further filter the results by entering keywords (top dashed-line box). Each record is linked to specific species page, miRNA page, target transcript page and the corresponding evidence page (Figure 5).
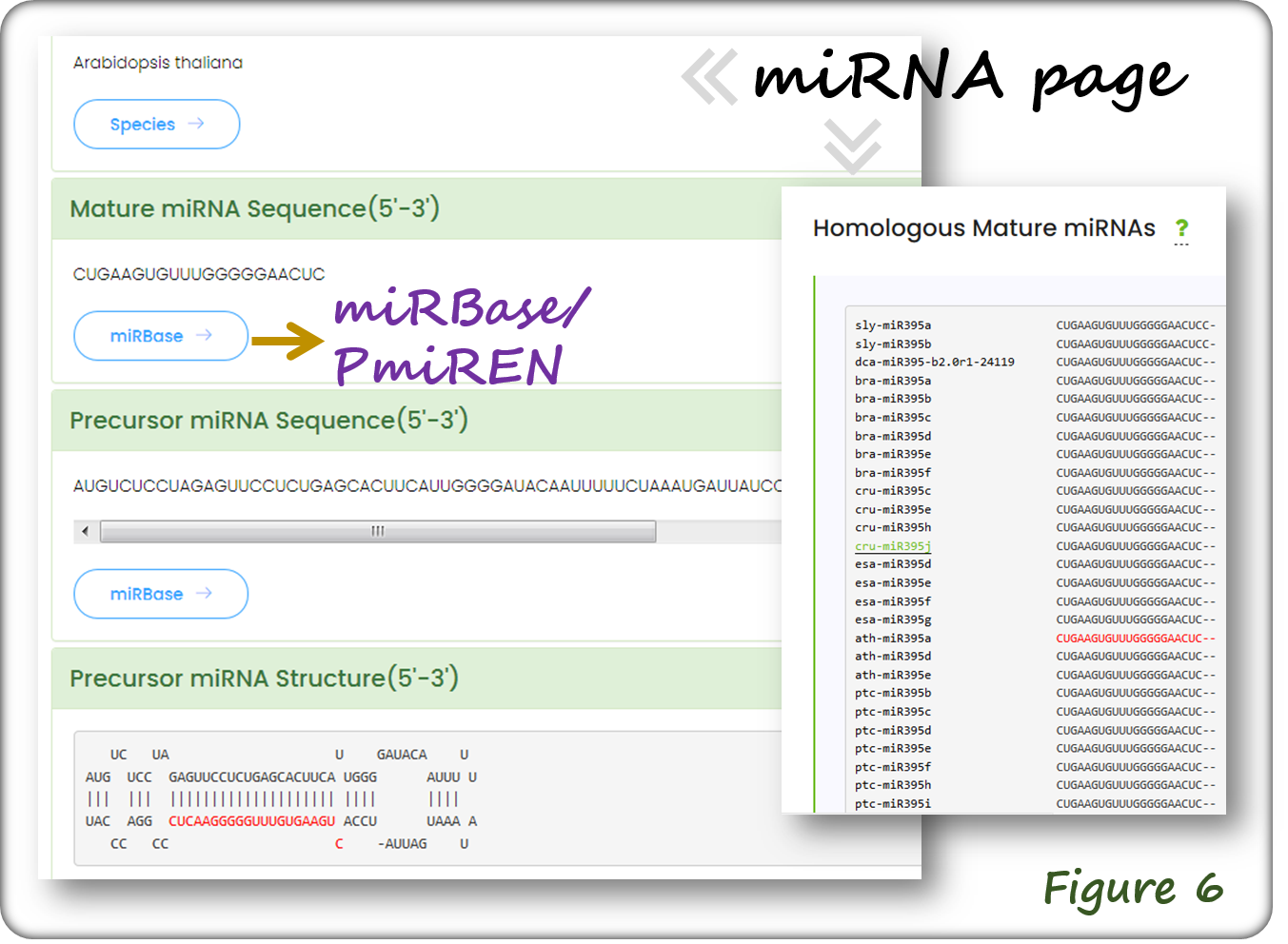
The miRNA webpage contains miRNA ID, mature and stem-loop structured pre-miRNA sequences as shown below(Figure 6). Each miRNA has external link to miRBase or PmiREN. Alignment of homologous miRNAs from various species is shown (Figure 6).
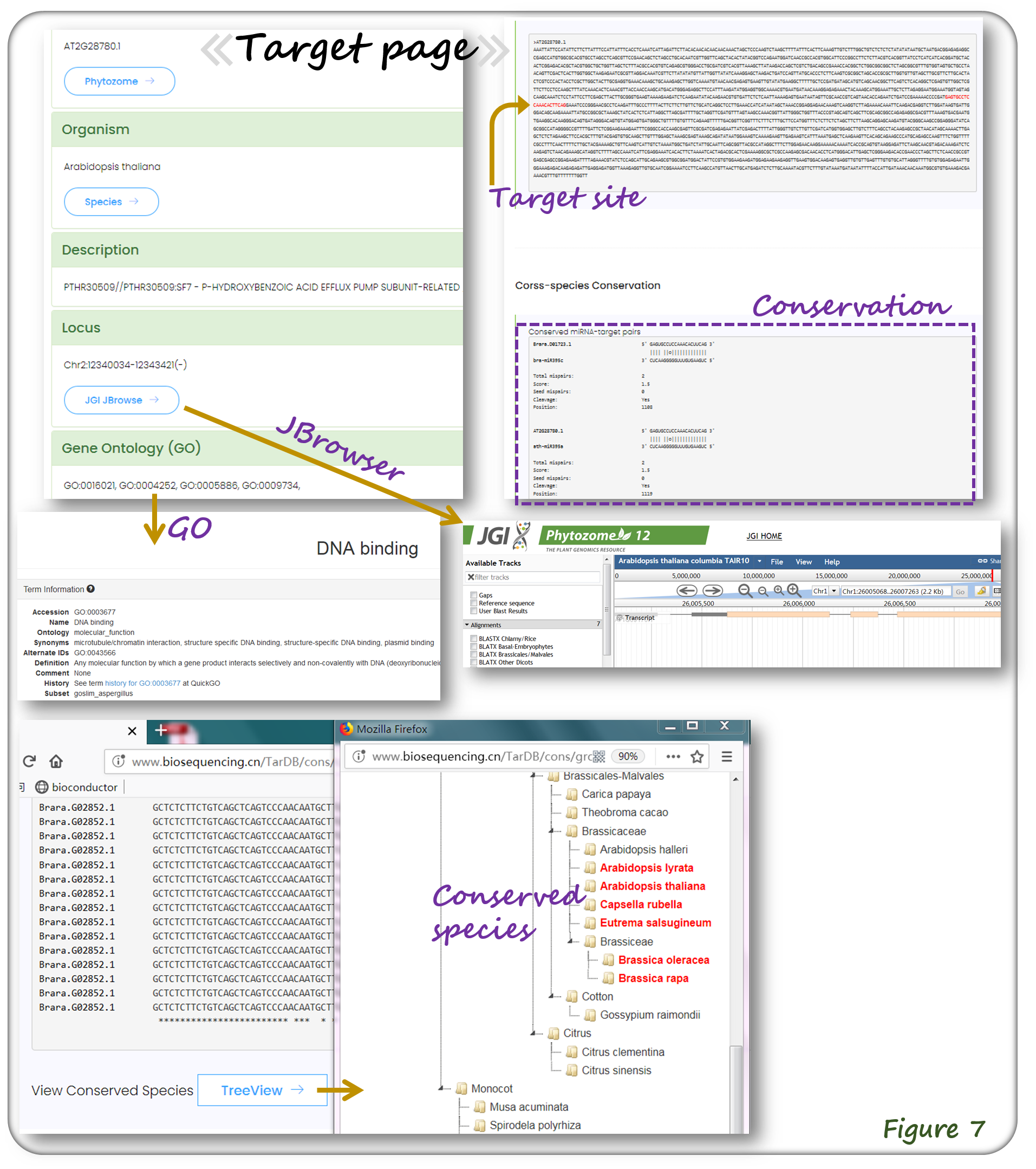
The target webpage contains transcript ID, sequence, annotation etc. which have hyperlinks to Genome Browser and Gene Ontology (GO) (Figure 7). The conservation of miRNA-target regulations in different species can also be viewed (Figure 7).
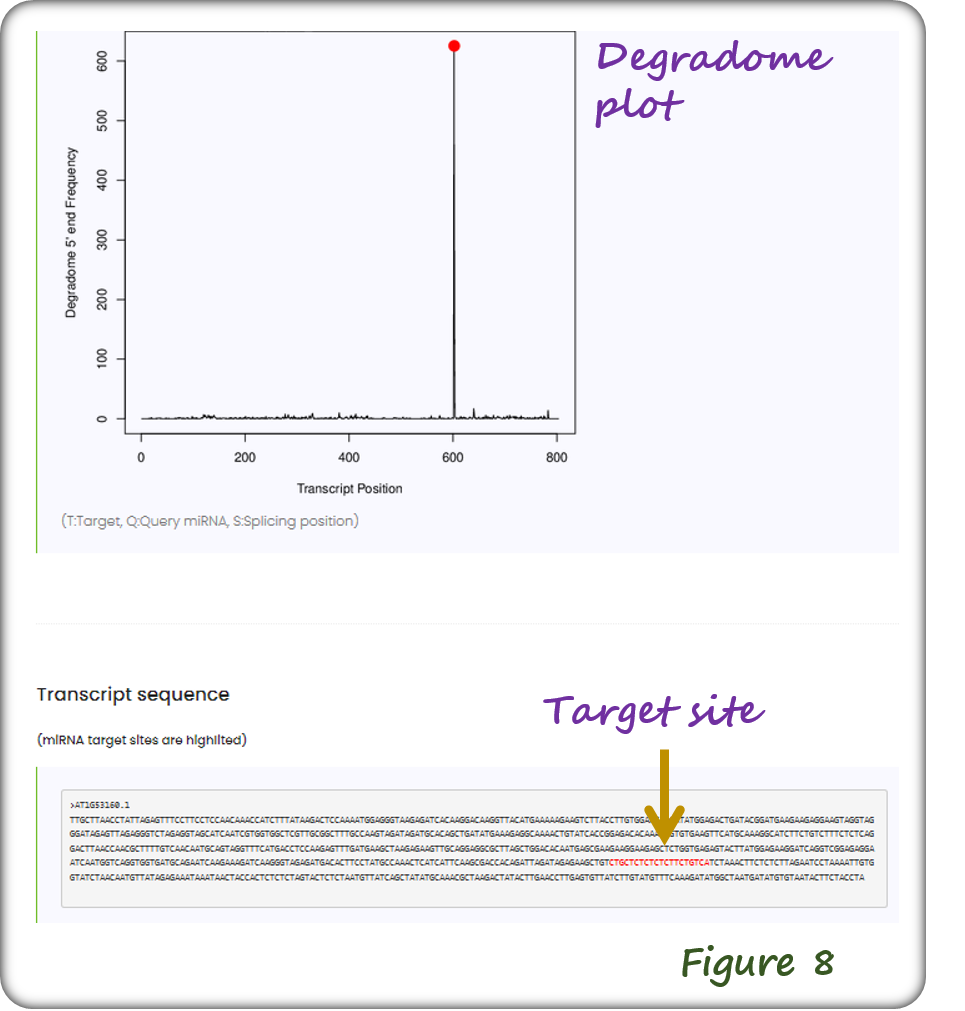
The webpage of degradome/PARE-seq supported miRNA targets includes degradome plot with miRNA target site highlighted (Figure 8).
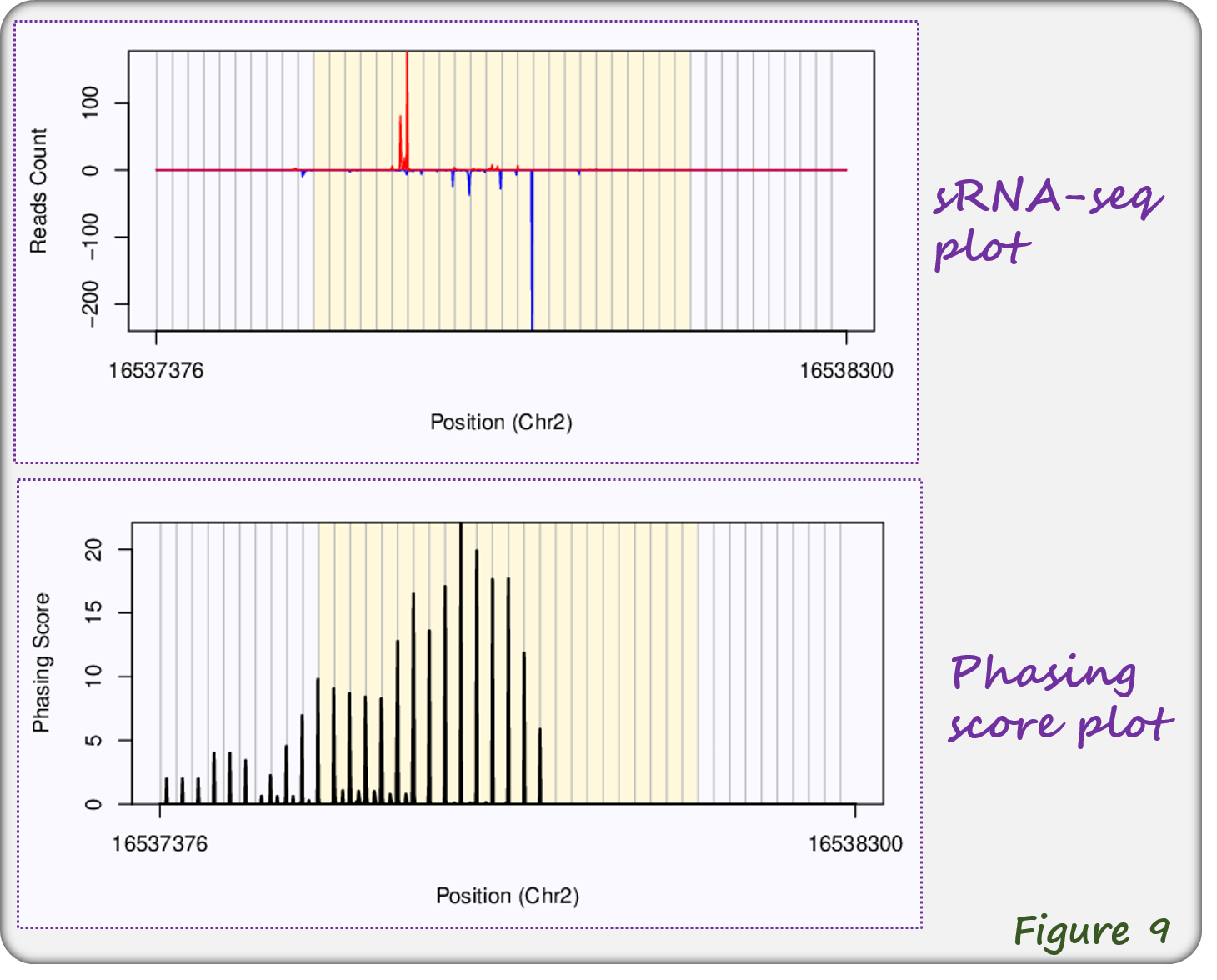
The webpage of miRNA-triggered phasiRNAs contains sRNA-seq profiles and phasing score plots (Watson and Crick strands are shown in red and blue colors, Figure 9).
4. Download
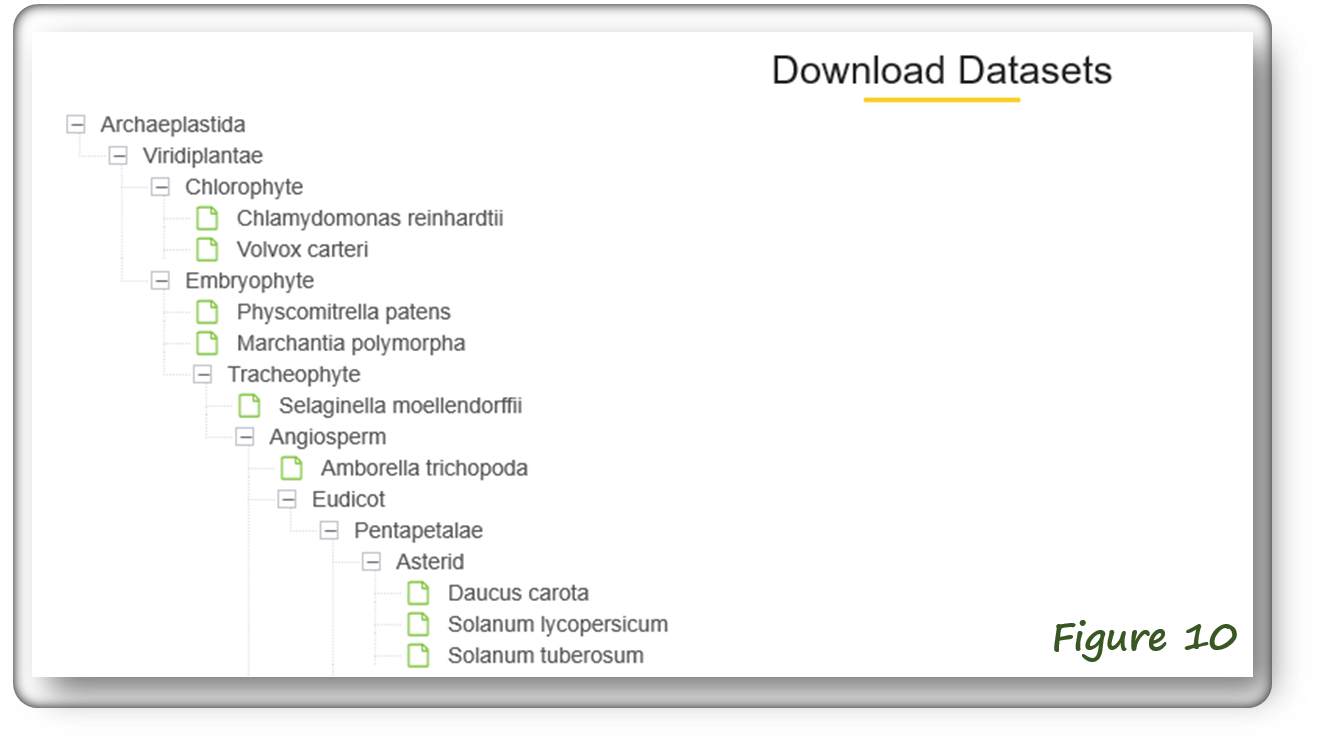
The Download webpage enables to download plant miRNA target data (in compressed file format) in various species. (Figure 10).
5. Guide
The Guide webpage presents details of our workflow and the codes in sequencing data analysis. It also contains useful links related to plant miRNA and target resources. (Figure 11).
6. Contact
The Contact webpage includes our email and work address for contacting. (Figure 12).
Related Codes
TarHunter (view code)is used to predict conserved miRNA targets with usage as follows (transcript data are stored under the phytozome12 directory, oth.fa contains miRNAs not deposited in miRBase)
TarHunter.pl -q mirlist -s spelist -b phytozome12/ -o out -f 5 -n oth.fa -G
CleaveLand4 (Addo-Quaye et al. Bioinformatics,2009; Mike Axtell lab) is used to analyze degradome/PARE-seq data in two steps. The 1st step runs one miRNA to generate degradome density file with usage as follows.
CleaveLand4.pl -e deg.fa -u mir1.fa -n trans.fa -o mir1 -c 2 -t
The 2nd step runs all miRNAs with the degradome density file as input with usage as follows.
CleaveLand4.pl -d dd_file -u mir.fa -n trans.fa -o out -c 2 -t
PHASIS (Kakrana et al. BioRxiv, 2017; Blake Meyers lab)
is a computational pipeline for phasiRNA discovery and annotation, as well as for identifying miRNA triggers.
PHASIS code is deposited on Github.
PhaseTank (Guo et al. Bioinformatics, 2015) is an easy-to-use pipeline for characterizing phasiRNAs and their regulatory networks.
PhaseTank code is available on Sourceforge.
FAQs
- What's the criteria in searching cross-species conserved miRNA targets?
- What's the criteria used in the degradome analysis?
- Could you explain different parameters in miRNA target prediction and degradome analysis?
- TarHunter Score: A scoring scheme represents the stringency of miRNA-target pairing. The fewer the score is, the more strict miRNA-target pairing is. In TarHunter scoring, mismatches or Indels are penalized 1 score. G-U wobbles are penalized 0.5 score. Penalties at positions 2-12 from miRNA 5' end are doubled. If positions 3-5 have mismatches, additional 1 score is penalized.
- TarHunter Homo mode: 50-bp upstream and downstream of the target sites are cross-species conserved (>70% sequence identity).
- Total Mispairs: All mismatches and Indels (insertions and deletions)
- Seed Region: Traditionally, miRNA 5' positions 2-7nt are recognized as seed region.
- Predicted Cleavage: Plant miRNA-target has high sequence complementarity. MiRNA induced cleavage of target RNA normally occurs precisely between positions 10 and 11.
- CleaveLand Category: Cat.0, Cat.1: maximum sequencing peak; Cat.2: above average sequencing depth
- Allen et al Score: Allen et al reported scoring system (Allen et al. 2005 Cell, 121:207-221 [PMID:15851028]; Fahlgren et al Methods Mol Biol 2010 [PMID: 19802588]) penalizes mismatches or bulges as 1 score. G-U wobbles are penalized 0.5. The penalties at positions 2-13 are doubled.
- CleaveLand P-value: P-value calculates the binomial probability of observations in a number of trials (see CleaveLand4 manual)
- How to identify miRNA-triggered phasiRNA loci?
Conserved miRNA targets are identified using TarHunter which requires orthologous miRNAs target homologous genes. Orthologous miRNAs are clustered by CD-HIT. TarHunter requires 50-nt upstream and downstream of target sites are conserved (i.e. >70% sequence identity in USEARCH).
Degradome analysis was performed using CleaveLand4 with the following criteria: (i) degradome category 0 or 1 or 2; (ii) Allen et al reported score ≤5. (iii) Binomial P-value ≤0.01
Phasing analysis was performed as previously described (De Paoli et al., 2009; Howell et al., 2007). Small RNA reads from sense and antisense strands were unified. Phasing score was calculated by the following formula.
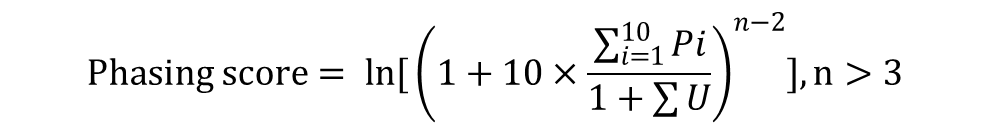
Hypergeometric test is used to further filter phasiRNA loci (Chen et al. PNAS,2007). The hypergeometric test statistically differentiates phased siRNAs from random siRNAs.
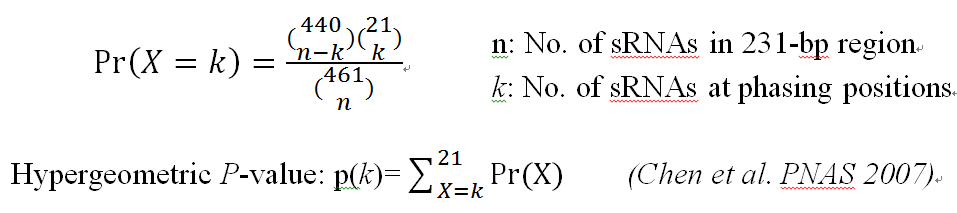
Finally, we manually examined sRNA-seq and phasing score profiles to obtain relatively high-confidence phasiRNA loci.
Related Links
Plant miRNA/sRNA Resources
- miRBASE
- PMIREN
- Penn State Plant sRNA Gene Server
- PMRD
- PmiRKB
- sRNA Data at Danforth Center
- sRNA Data at Blake Meyers Lab
- CSRDB
- Pln24NT
- ASRD
- PhasiRNAnalyzer